The human heart is an amazing machine that lasts for a century without fail. One of the key methods of measuring cardiac function is to calculate its ejection fraction, which is the percentage of stroke volume as a percentage of ventricular end-diastolic volume. The first step in measuring this indicator relies on the segmentation of the heart image ventricles.
Problem DescriptionDevelop a system that can automatically segment the right ventricle in a cardiac magnetic resonance imaging (MRI) dataset image. So far, this has mainly been handled by classical image processing methods. And modern deep learning technology is likely to provide a more reliable and more automated solution.
The three winners of the Left Ventricular Segmentation Challenge sponsored by Kaggle in 2016 adopted a deep learning solution. However, dividing the right ventricle (RV) is more challenging because:
There are trabeculae in the cavity similar to myocardial angiography; RV complex crescent; difficulty in segmenting apical image slices; there is considerable variation in the shape and intensity of the individual between individuals, especially in different disease cases between.
Regardless of medical terminology, it is more difficult to identify RV. The left ventricle is a thick-walled ring, and the right ventricle is an irregularly shaped object with thin walls that sometimes mix with the surrounding tissue. This is a hand-drawn outline of the inner and outer walls of the right ventricle (endocardium and epicardium) of the MRI snapshot:
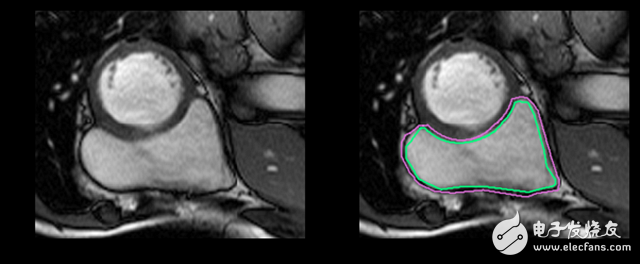
This is an easy example of segmentation. This one is more difficult:
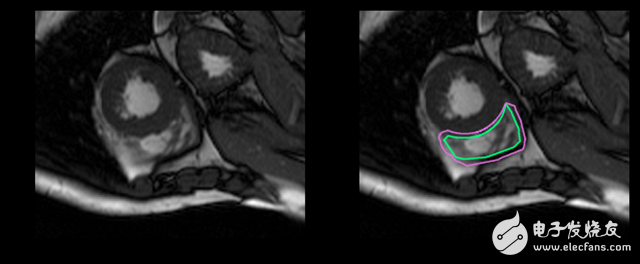
And this is a complete challenge for untrained eyes:
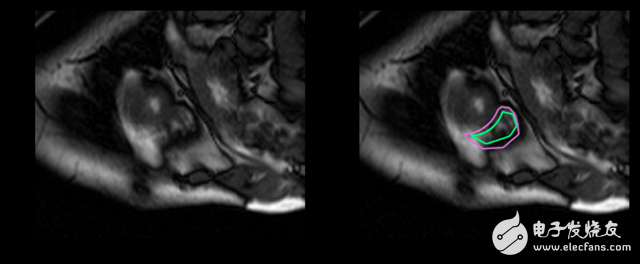
In fact, compared to the left ventricle, the doctor takes twice as long to determine the volume of the right ventricle and produce a result. The goal of this work is to establish a highly accurate deep learning model for right ventricular auto segmentation. The output of the model is a *split mask*, a pixel-by-pixel mask that indicates whether a pixel is part of the right ventricle or just a background.
data setFor the current problem, the biggest challenge for deep learning is that the data set is too small. The data set (accessible here) contained only 243 physician-division MRI images from 16 patients. There are also 3,697 unlabeled images, which may be useful for unsupervised or semi-supervised techniques, but I put this aside because it is a supervised learning problem. The image size is 216 & TImes; 256 pixels.
Because the data set is too small, one might suspect that it cannot be generalized to an image that has not yet been seen! But unfortunately, medically labeled data is very expensive and difficult to obtain. The standard procedure for processing data sets is to apply affine transformations to the image: random rotation, translation, scaling, and clipping. In addition, I achieved elastic deformation, which is to stretch and compress the local area of ​​the image.
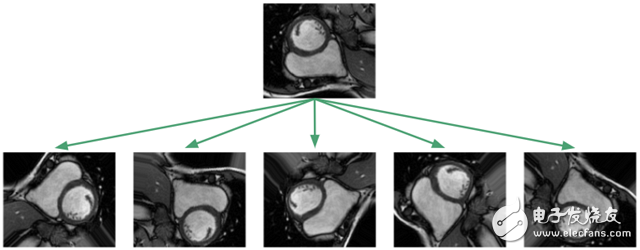
The purpose of applying this image enhancement algorithm is to prevent the neural network from memorizing only the training examples and forcing them to learn that the RV is a solid, crescent-shaped, arbitrarily oriented object. In the training framework I implemented, I will apply the image transformation algorithm to the dataset at any time, so that the neural network will see a new random transformation every time we train.
Since most of the pixels belong to the background, there is an imbalance between the various categories. If the pixel intensity is normalized between 0 and 1, then only 5% of the pixels in the entire data set belong to a portion of the RV cavity.
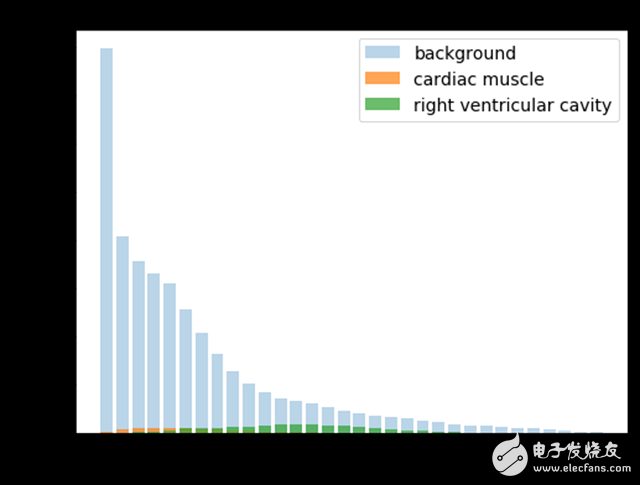
When creating the loss function, I tried a re-weighting scheme to balance the distribution of the categories, but eventually found that the unweighted averaging algorithm performed best.
In training, 20% of the images were taken out for use as a validation set. The organizers of the RV Segmentation Challenge have a separate test set consisting of 514 MRI images from another 32 patients. The predicted profile I submitted is the final evaluation using this test set.
There is also a need to have a way to quantify the performance of the model on the dataset. The organizers of the RV Split Challenge chose to use the Dyce coefficient. The model outputs a mask *X* to describe the RV, and the Daysian coefficient compares *X* with the mask *Y* created by the physician in the following ways:
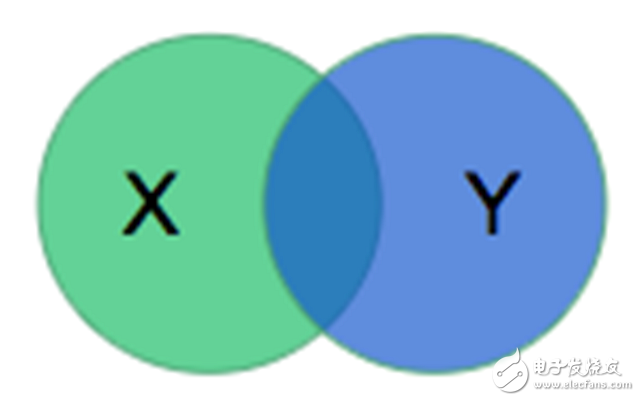

The calculated value is twice the ratio of the intersection area to the sum of the two areas. For disjoint regions, the value is 0; if the two regions are identical, the value is 1.
Let's take a look at the model architecture.
U-net: baseline modelSince we only have four weeks to complete the project, I want to implement a baseline model and get it running. I chose the u-net model proposed by Ronneberger, Fischer, and Brox because it was successful in biomedical segmentation projects, and its authors were able to use positive image enhancement and image-by-image re-weighting algorithms based only on * 30 pictures* to train the network.
The u-net architecture consists of a shrink path that folds the image into a set of advanced features, followed by an extension path that uses the feature information to construct a pixel split mask. The unique thing about u-net is its "copy and merge" connections, which pass information from the early feature map to the subsequent parts of the network that builds the split mask. The authors point out that these connections allow the network to incorporate high-level features and pixel details at the same time.
The architecture we use is as follows:
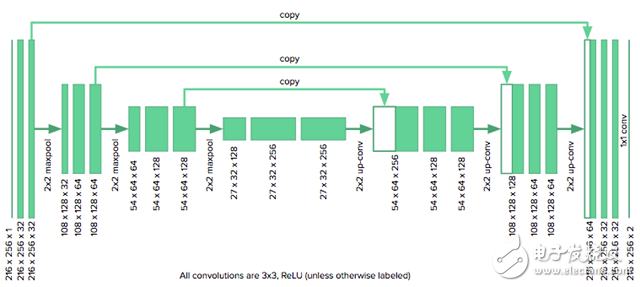
Since our image size is half the size originally considered by u-net authors, we need to reduce the number of downsampled layers in the original model from four to three to fit the grid. We also need to fill the convolution with zeros to keep the size of the image unchanged. The model is implemented using Keras.
Without image enhancement, the U-net's Dyce coefficient on the training data set can reach 0.99 (0.01), which means that the model has enough power to capture the complexity of the RV segmentation problem. However, verifying the Dyce coefficient is 0.79 (0.24), so u-net is too strong. Image enhancement improves generalization and increases verification accuracy to 0.82 (0.23) at the cost of reducing training accuracy to 0.91 (0.06).
How can we further reduce the gap between training and verification? As Andrew Ng described in this great conversation, we can use more data (which is unlikely), regularization (dropout and batch normalization have no effect), or try a new model architecture.
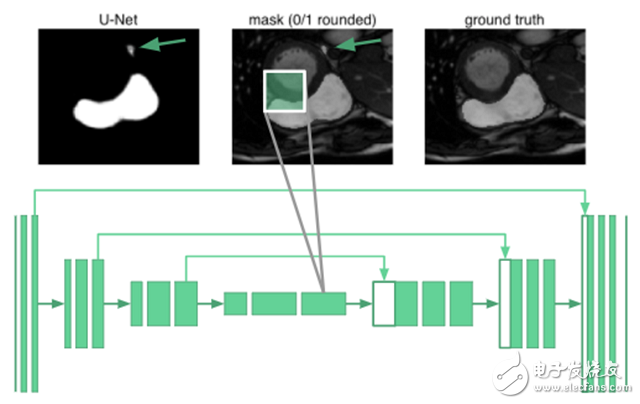
Expanding U-net: Global ResilienceTo segment an organ image, you need to know the relevant knowledge of the arrangement between organs. It turns out that even the deepest neurons in the u-net are only 68 & TImes; 68 pixels of the receptive field. No part of the network can "see" the entire image. The network does not know that humans have only one right ventricle. For example, in the image below, the place marked by the arrow is misclassified:
We use expansion convolution to increase the receptive field of the network.
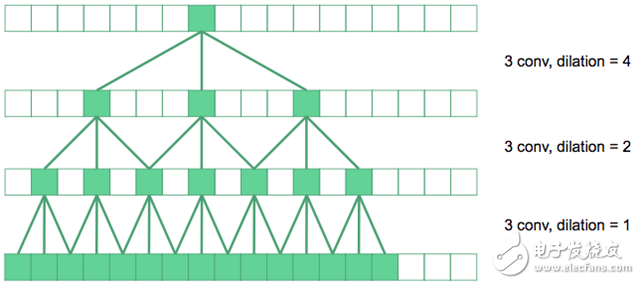
In the figure above, the underlying convolution is the regular 3&TImes;3 convolution. On the next level, we have expanded the convolution by a factor of two, so their effective receptive field in the original image is 7&TImes;7. If the top convolution is expanded by a factor of four, a 15 x 15 receptive field can be produced. And so on.
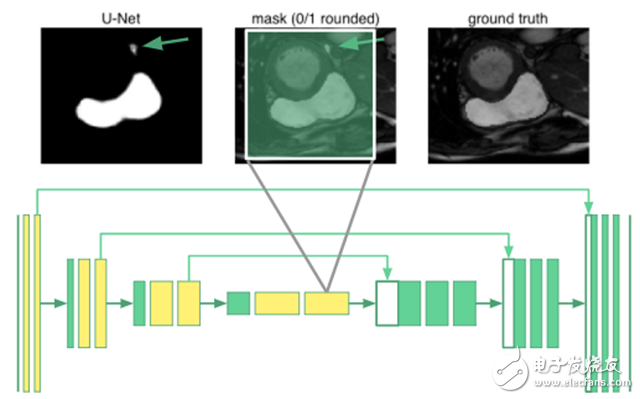
In principle, the yellow-marked convolutional layer is replaced by an expanded convolution in the u-net. The innermost neurons now have a receptive field that covers the entire input image. I call it "expansion u-net."
In terms of quantity, the expansion of u-net did improve the effect, reaching a 0.85 (0.19) verification Dyce score while maintaining a training effect of 0.92 (0.08)!
Expand DenseNet: one-off multiple scalesThis inspiration comes from the tensor network in physics, and I decided to try a new image segmentation architecture, which I call "expanding DenseNet." It combines the two concepts of expanding convolution and DenseNet, which can greatly reduce the depth and parameters of the network.
For segmentation, we need global context and information from multiple scales to produce a pixel-level mask. If we rely entirely on expanding convolution to produce a global context, rather than downsampling to make the image smaller? Now that all convolutional layers are the same size, we can apply the key idea of ​​the DenseNet architecture and use a "copy and merge" connection between all layers. The results of expanding DenseNet are shown below:
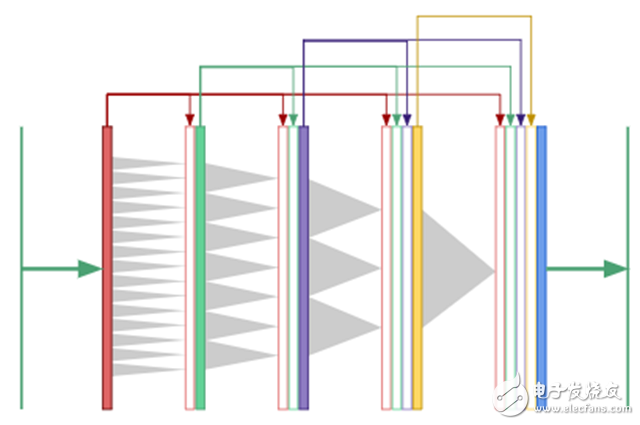
In DenseNet, the output of the first convolutional layer is fed as input to all subsequent layers, as are the second and third layers.
The expansion of DenseNet performed well, with a 0.87 (0.15) Dyce score on the verification set, with a training accuracy of 0.91 (0.10) while maintaining extremely high parameter efficiency!
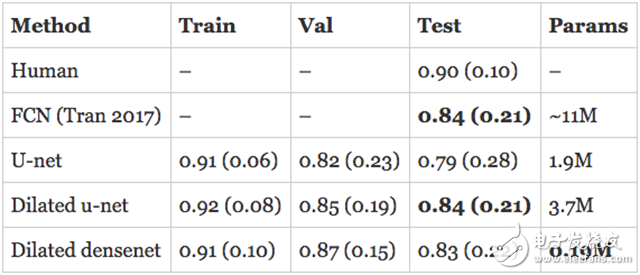
resultThe assessment of human RV segmentation indicates the direction of how to evaluate the performance of the model. The researchers estimate that the Dyce score for humans completing the RV segmentation task is 0.90 (0.10). The published model described above is a Full Convolutional Network (FCN) with an accuracy of 0.84 (0.21) on the test set.
The model I developed has surpassed the latest state of the art in the validation set and is approaching human performance! However, the real assessment is to evaluate the performance of the model on the test set. In addition, the numbers quoted above are for the endocardium, so what is the performance of the epicardium? I trained a separate model on the epicardium and submitted the subdivision to the organizer in hopes of getting the best results.
Here are the results, first the endocardium:
This is the epicardium:
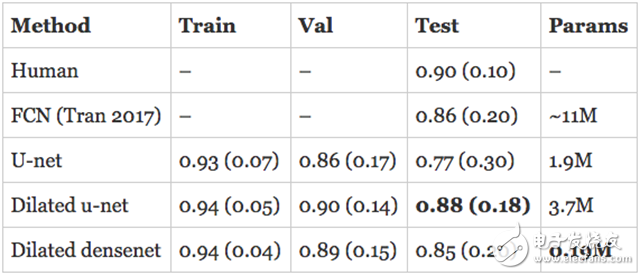
The expansion of u-net is comparable to the state of the art in the endocardium and exceeds its performance on the epicardium. The expansion of DenseNet followed closely with only 190K parameters.
to sum upThe performance of the deep learning model sometimes seems magical, but it is the result of careful design. Even with a small data set, a carefully selected data enhancement scheme can make the deep learning model more general.
Based on these ideas, a state-of-the-art model was created to segment the right ventricle in cardiac MRI. I am very happy to see that the expansion of DenseNet can be successfully run on other image segmentation reviews.
Memory-efficient expansion of DenseNet: A densely connected network has a big drawback, it requires a lot of memory. The implementation of TensorFlow is different, it limits us to a 16GB GPU and a batch of 8 layers with 3 images. If you switch to the previously efficient implementation of memory, you can create a deeper architecture.
Shenzhen ChengRong Technology Co.,Ltd. , https://www.laptopstandsuppliers.com